Our blog
We address a broad spectrum of topics to cater to your questions and interests comprehensively. From industry trends to practical tips, our content is designed to provide valuable insights and answers to meet your needs.
Discover moreResources
We address a broad spectrum of topics to cater to your questions and interests comprehensively. From industry trends to practical tips, our content is designed to provide valuable insights and answers to meet your needs.
Discover moreBlog

Towards Integrated Oncology Diagnosis: Artificial Intelligence as the Backbone
Published on 10/12/2025
2 min
Blog

Behind the Scenes of Building an AI Algorithm in Digital Pathology
Published on 30/07/2025
4 min
Blog
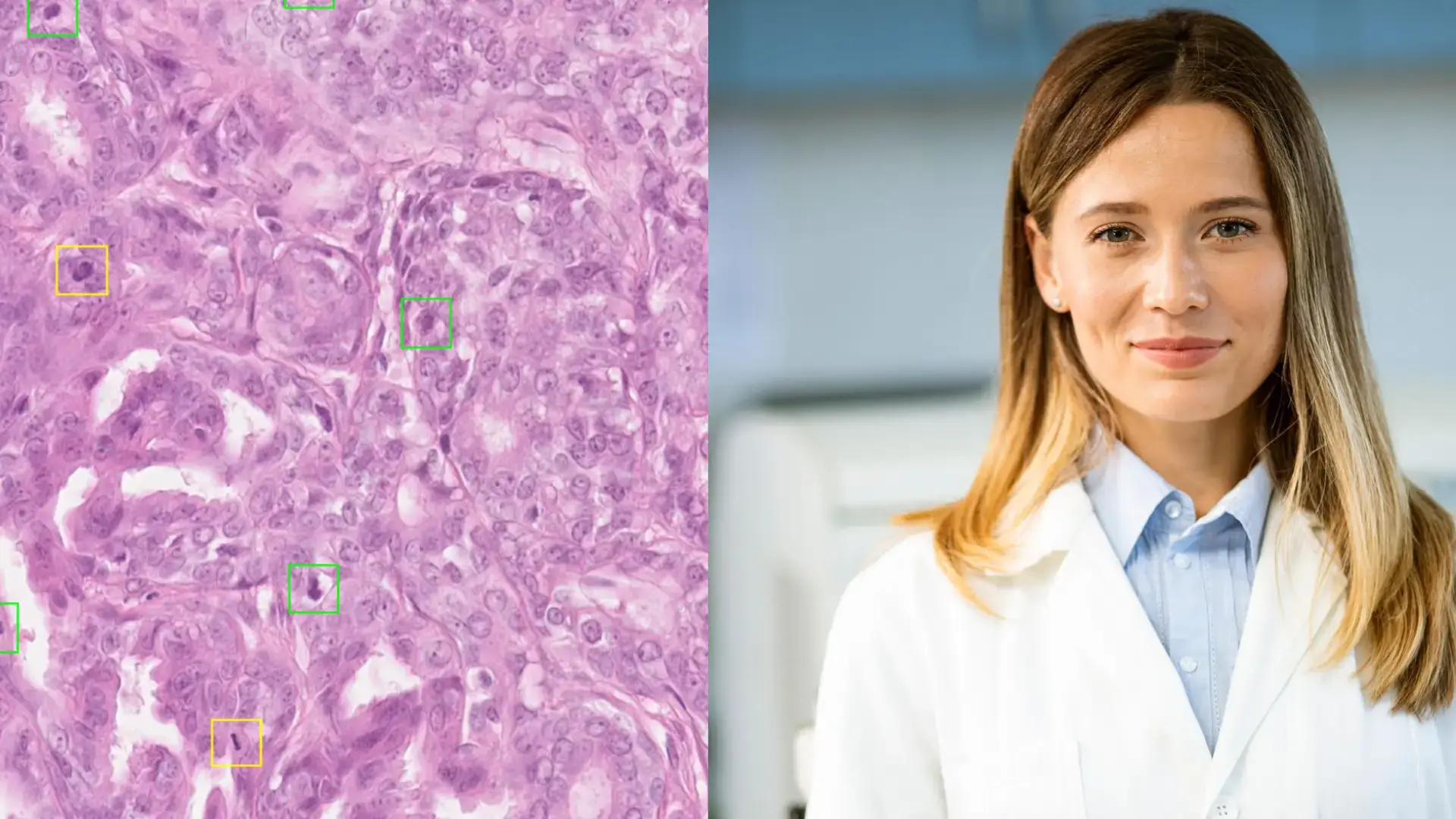
Transforming Pathology: How AI Brings Precision and Faster Access to Care
Published on 29/07/2025
3 min
Events

See the event
USCAP 2026
Join Tribun Health at USCAP 2026 in San Antonio.
From 23/03/2026 to 25/03/2026
San Antonio
315
Curious about upcoming conferences, webinars, or events? Stay in the loop and discover what's on the horizon for the next few weeks. Explore our calendar to see the exciting lineup of upcoming opportunities to engage, learn, and connect within our community.
Discover our eventsDive into our video section to experience a dynamic array of content, including client testimonials, insightful interviews, post-conference discussions, webinars and engaging team stories.
DISCOVER OUR VIDEOS
APHP testimonial: Use of RlapsRisk® BC and CaloPix®.
Published on 16/07/2024
4 min

APHP testimonial: Use of RlapsRisk® BC and CaloPix®.
Find out how RlapsRisk® BC uses digitised slides and the CaloPix® IMS to predict the risk of recurrence at diagnosis. The video also provides an overview of the development of the tool, highlighting the importance of feedback from pathologists and oncologists to refine and perfect the model.
Published on 16/07/2024
4 min
Interview on the growth of Digital Pathology with Jean-François Pomerol at the HBI
Published on 03/07/2024
3 min

Interview on the growth of Digital Pathology with Jean-François Pomerol at the HBI
Jean-François Pomerol, CEO at Tribun Health is thrilled to have been invited as a speaker at the Healthcare Business International (HBI) event. Jean-François had the honor of sharing his insights in an exclusive interview with Chris O'Donnell, Head of Content & Editorial HBI. He discussed digital pathology, its deployment, its integration into the pathologist's workflow, the use of AI, and the challenges and opportunities of the sector.
Published on 03/07/2024
3 min
Explore our 'News' section to stay updated on our latest announcements and industry news. Don't miss out on important updates and developments within our company and the broader industry.
Discover our newsExplore our 'Customer Testimonials' section to discover authentic feedback from our clients. Delve into the successes, challenges overcome, and results achieved through our solutions. Be inspired by the success stories and satisfaction of our customers.
Discover our customers stories
See the testimony

Published on 17/07/2024
AP-HP Testimonial : The use of RlapsRisk® BC and CaloPix®
Recording by
Orion Films
See the testimony
Explore this dedicated section for a comprehensive array of resources including our latest scientific articles, thought-provoking abstracts, informative posters, in-depth white papers, and insightful articles featured in specialized press magazines.
Discover more
N°1
DigPath Platform I 3x Best in KLAS Winner
+100
Customers, including diagnostic & pharma labs
+50
Specialists forging reliable solutions

Discover Tribun Health's end-to-end platform, comprising all the modules we offer: MacroCam, CaloPix Archive, AI Apps, CaloPix® and TeleSlide. Together, they meet the end-to-end digitization needs of pathology departments, and integrate seamlessly into pathologists' workflows. At the heart of the digital pathology workflow is our Image Management System (IMS), CaloPix®.
